Sarai and Alex recently won best poster awards for their posters on engineering STARs and RNA circuits at the 2015 Fall SynBERC retreat! Congratulations team!
Author Archives: jblucks
Lucks Lab helps ‘Build with Biology’
We recently helped out with a new public engagement activity called ‘Building with Biology’! Sponsored by the National Science Foundation and based at the Museum of Science in Boston, Building with Biology is a program designed to promote conversations between the public and scientists about synthetic biology. As part of the project, we have beenContinue reading “Lucks Lab helps ‘Build with Biology’”
Our in-cell SHAPE-Seq paper is out!
Our method for characterizing the cellular structure and function of RNA regulators is out in Nucleic Acids Research. The paper outlines our method for applying SHAPE-Seq to characterize RNA structures within E. coli cells by directly applying the 1M7 SHAPE reagent to cell culture. Since we are very interested in RNAs that regulate gene expression,Continue reading “Our in-cell SHAPE-Seq paper is out!”
Kyle Becomes an Inaugural Fleming Fellow!
Kyle has been selected as an inaugural Fleming Graduate Scholar in the School of Chemical and Biomolecular Engineering. The Fleming Scholarships are due to a generous gift from Sam and Nancy Fleming to support the most outstanding PhD students pursuing research in biomolecular engineering at Cornell. Congratulations Kyle!
Welcome New Lucks Lab Members!
We have an influx of new Lucks Lab members! Welcome Angela, Aron, Jane, Yan and Raashed!
Another Great Year at CSHL SynBio
We had another fantastic year at the Cold Spring Harbor Laboratory Synthetic Biology summer course! Led by James (universally voted a Rock STAR scientist by everyone at the course), we successfully delivered a thrilling module on characterizing genetic circuitry using cell-free TX-TL reactions. Actually I can’t thank James enough for all the hard work both beforeContinue reading “Another Great Year at CSHL SynBio”
A Renaissance in RNA Synthetic Biology
A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future is out in Current Opinion in Chemical Biology! James, Kyle and Melissa did a fantastic job making our case for why we think we are in a renaissance in RNA synthetic biology right now. In the review, we go over the latestContinue reading “A Renaissance in RNA Synthetic Biology”
Improving STARs with Rational RNA Engineering Strategies
Our paper on improving Small Transcription Activating RNAs with Rational RNA Engineering Strategies is out in Biotechnology and Bioengineering! In this paper, Sarai, James, Sitara and Rebecca outline several different strategies for improving the fold activation of weak STARs, including promoter tuning and RNA stabilization strategies. We hope this helps others use STARs in theirContinue reading “Improving STARs with Rational RNA Engineering Strategies”
Our RNA Circuitry Modeling Paper is Out!
Our paper on Generating effective models and parameters for RNA genetic circuits is out in ACS Synthetic Biology. In this paper, Chelsea developed a simplified ODE model to model the dynamics of RNA genetic circuits. These ‘effective’ models essentially course grain a lot of the biochemical details of gene expression into simple mass-action kinetics thatContinue reading “Our RNA Circuitry Modeling Paper is Out!”
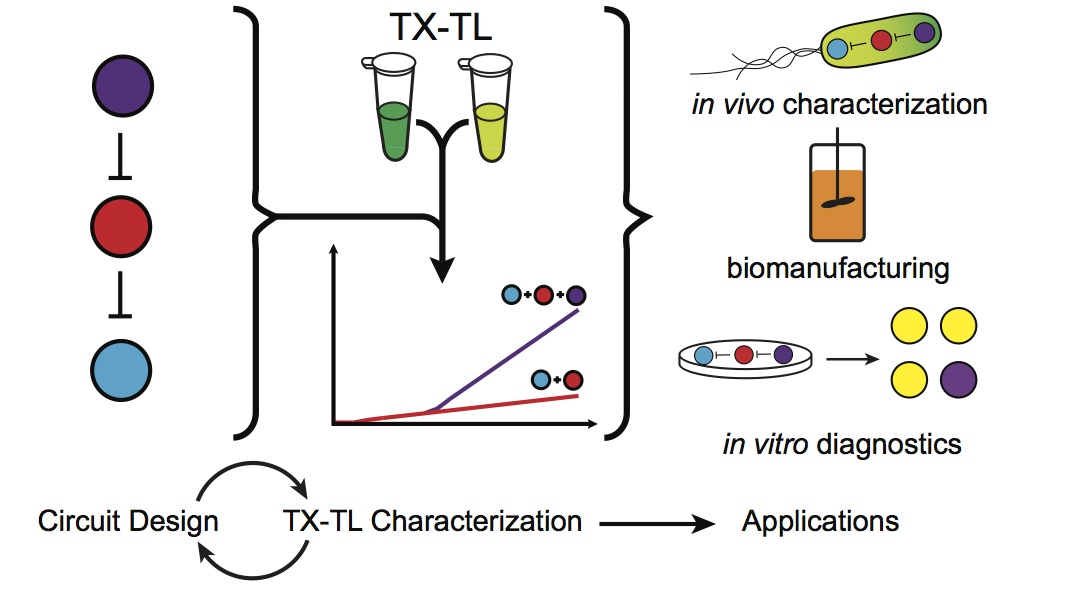
Our TX-TL Methods Paper is Out!
We were recently invited to prepare a paper for the journal Methods about using cell-free transcription-translation reactions (TX-TL) to characterize genetic circuits. We immediately joined forces with Vincent Noireaux’s and Richard Murray’s groups and had a lot of fun putting this together! What started out as a brief intro to the basics behind TX-TL turned intoContinue reading “Our TX-TL Methods Paper is Out!”